DiseasedCNN-Lite: Leveraging Transfer Learning and Knowledge Distallation for Leaf Disease Classification
Abstract
Identification of diseased crops through the symptoms displayed on their leaves can help farmers manage large-scale operations. Integrating deep learning with robotics provides a promising way for farmers to maintain their crops in an efficient and cost-effective manner. However, deep learning models are often too large and computationally complex to be deployed in agricultural robots. Previous research has been conducted on small datasets and has produced models that can identify diseases for a specific species of crop. Drawing from previous research on the study of apple leaves and associated diseases, along with studies on knowledge distillation, this work proposes a lightweight model that can classify 17 common diseases spanning 14 different crop species.
Motivation and Original Paper
Agricultural robotics is a fast-growing area of research—in particular, many researchers are focusing on using automation to perform tedious everyday tasks such as weeding, scouting, and harvesting. Properly trained robots would be less prone to missing important information when working fields, and would not fall victim to dangerous environmental factors such as extreme heat, pesticides, and hazardous machinery. As robotics has become a more prominent field in engineering, interest in agricultural robotics has spiked, and deep learning has been a heavily researched area relating to this topic. However, as interest in this specific area of robotics has increased, the requirements for a practical machine learning model have become narrower. Any model created must be highly accurate, generalizable to real-world data, and require as little memory and computational power as possible, such that a robot’s ability to perform tasks remains more efficient than a human’s.
Image processing and classification of diseased leaves has been researched before, and a majority of studies are specific to one crop (apples and tomatoes are by far the most common). Many different machine learning architectures have been attempted—some deep models, some not—and most have been able to achieve test accuracies upwards of 90%-95%. These studies focused on one crop species and had relatively small datasets of a few thousand training images creating a lack of versatility.
DeepCNN is a convolutional neural network that can identify three different apple leaf diseases, as well as a healthy leaf. The study laid the foundation for our work on disease detection. After training for 1000 epochs, DeepCNN achieved an accuracy of 98% on a dataset consisting of 3,171 images (2,228 training, 634 validation, 319 testing). To lay the foundation for our model, we attempted to reproduce the final results. The study provided the dataset used and outlined the exact architecture of DeepCNN, including specific dimensions for each layer; however, specifics for the data augmentation performed were not provided.
Our Extensions
In our attempt to reproduce these results, we discovered that DeepCNN would drastically overfit to the data when trained for 1,000 epochs. Instead, we achieved 96% testing accuracy after 250 epochs of training, and training for any longer would lead to a decrease in overall model performance. We theorize that this discrepancy could be a result of the differing data augmentation. Since the operations performed on the images were not detailed, we researched and used relatively common operations (horizontal and vertical flips, shearing, scaling, shifting) and applied them to each training image with a 40% probability. Data augmentation is used to prevent overfitting, but its misuse can harm model performance and actually increase the chance of overfitting. Therefore, we believe that our data augmentation was substantially different, and led to the drastic change in accuracy as the number of training epochs passed 250.
One limiting factor of the DeepCNN network proposed was that the dataset used only contained four classes on a single species. Therefore, we first aimed to build a network that could identify many more species and diseases at a similar rate. To do this, we used the Plant Village dataset. This dataset contains 54,305 images spanning 14 different species of crop and 17 common diseases, amounting to 38 separate classes. We split the dataset into training, testing, and validation using the standard 70-20-10 split.
As a preemptive measure to prevent overfitting, we probabilistically applied different augmentation operations to each training image. Given that there were only approximately 1,000 images per class in the Plant Village training set, we determined that data augmentation would be our most effective tool against overfitting. Our data augmentation consisted of shearing, scaling, horizontal and vertical flipping, and elastic distortion.
To determine which pretrained model to use as a backbone for transfer learning, we benchmarked the performance of seven commonly used models against the Plant Village dataset. This process included adding one fully connected layer to the end of each pretrained model, and then training for 20 epochs. EfficientNet-B0 and ResNet50 performed best. Both reached a loss below 3.0 and a validation accuracy above 80%. After this benchmarking process, we trained DiseasedCNN for three epochs with both EfficientNet-B0 and ResNet50 to determine which model meshed best with our fine-tuning layers. We determined that ResNet50’s feature extraction was better suited for our needs, as EfficientNet-B0 achieved a test accuracy of 61.51% and ResNet50 achieved a test accuracy of nearly 75%. Therefore, we decided to use ResNet50 as our pretrained backbone for DiseasedCNN.
DiseasedCNN
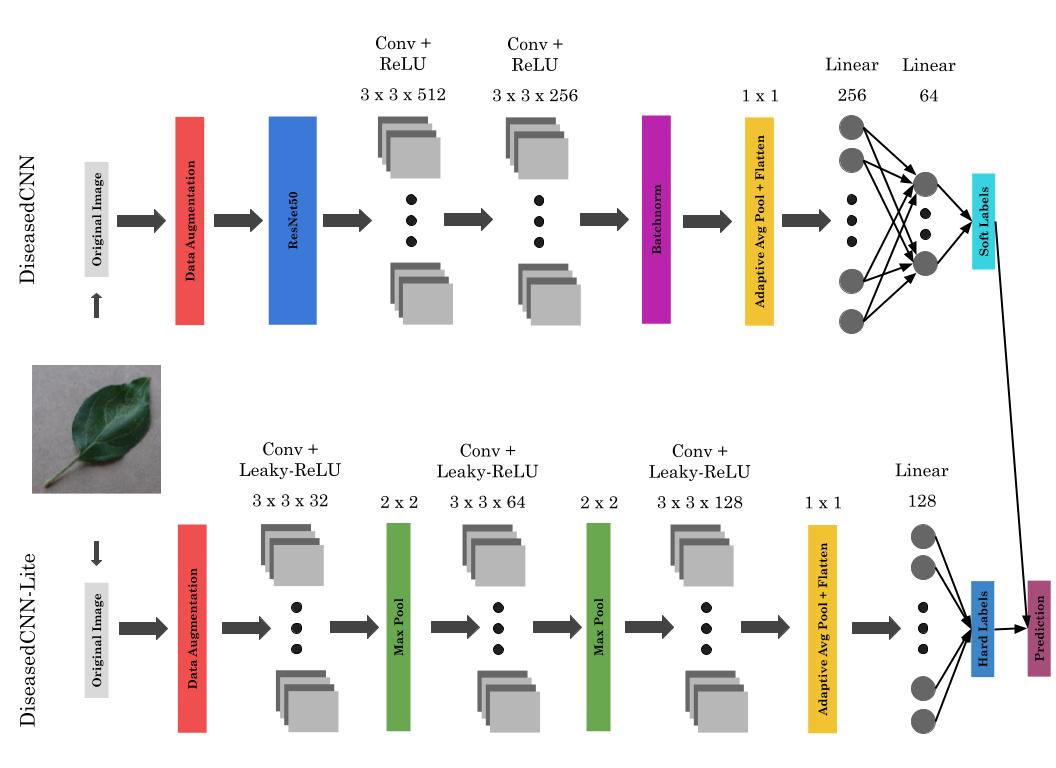
DiseasedCNN is the powerful model that we built to serve as a teacher during the knowledge distillation process. It relies on ResNet50’s feature extraction to provide a basis for its classification abilities, and utilizes fine-tuning layers to calibrate its prediction to our dataset. It also employs a rectified linear unit (ReLU) activation function to provide nonlinearity, and softmax is used on the output to produce a probabilistic prediction.
The foundation of DiseasedCNN is a ResNet50 backbone with the last three layers (adaptive average pooling, flattening, and fully connected) removed. This allows us to feed the output of the network into our fine-tuning layers, while maintaining the spatial information that the network has extracted.
The fine-tuning layer architecture is as follows: two convolutional layers with ReLU activations on the outputs, one batchnorm layer and one adaptive average pooling layer, and two fully connected layers, with ReLU activation after the first layer and softmax after the second.
Results
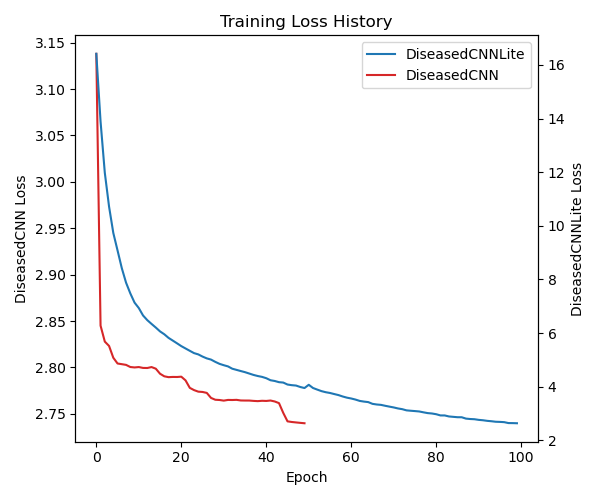
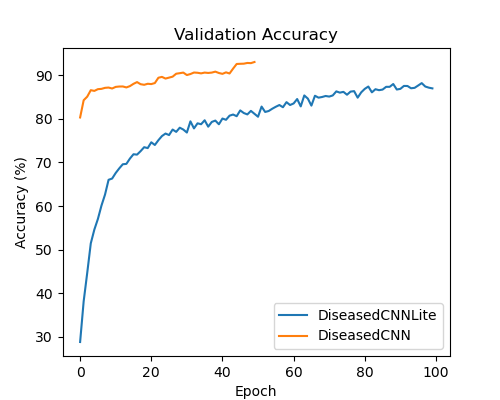
DiseasedCNN trained for 50 epochs on the Plant Village dataset, with model validation performed every 13 batches. During training, we used an Adam optimizer with a learning rate of 0.0005, a beta_1 value of 0.9, and a beta_2 value of 0.999. In addition, we used an exponential learning rate scheduler with a gamma factor of 0.9. Our loss function for DiseasedCNN’s training was the standard PyTorch cross-entropy loss function. The model was trained for approximately two hours, and while the average epoch loss remained relatively high, the model reached a final validation accuracy of 93%.
DiseasedCNN’s inference time is relatively high compared to other models of the same size. We would like to see this value within the range of 10 to 30 milliseconds. This is particularly important for any integration with agricultural robotics, because a longer inference time means that it takes more energy and computational power to run inference. Maximizing battery life is a major aspect of agricultural robotics. Furthermore, DiseasedCNN is too large for many agricultural robots and drones. This is largely due to its ResNet50 backbone, which provides the necessary feature extraction and enables such high classification accuracy. Overall, DiseasedCNN is a good start, but in order to be a practical solution in the field of optimized agricultural robotics, DiseasedCNN would need to see a drastic reduction in size and inference time.
DiseasedCNN-Lite
DiseasedCNN-Lite is a much smaller model that we built to serve as a student during the knowledge distillation process. Instead of relying on a large, pretrained backbone such as ResNet50 to perform feature extraction, it extracts a small number of foundational features from a given image, while focusing on learning to predict the output of its teacher, DiseasedCNN. Since this model is much smaller than DiseasedCNN, we opted to use a Leaky ReLU activation function to prevent any gradients from vanishing during the training process.
Results
DiseasedCNN-Lite trained for 100 epochs on the Plant Village dataset, with model validation performed at the end of each epoch. During training, we used many of the same hyperparameters as DiseasedCNN. We used an Adam optimizer with a learning rate of 0.0005, a beta_1 value of 0.9, and a beta_2 value of 0.999. We also used a learning rate scheduler to help optimize the training of the student model, with a gamma factor of 0.9. However, for DiseasedCNN-Lite we added a weight decay factor of 0.00001 to avoid large weights and prevent overfitting. Furthermore, DiseasedCNN-Lite trained with a T value of 4, as we chose to soften the teacher model’s probability distribution, and an alpha value of 0.7, putting more emphasis on hard loss than soft loss.
DiseasedCNN-Lite’s inference time was much faster than DiseasedCNN’s, and will likely require significantly less energy and computational power to run on an agricultural robot. In addition to this, DiseasedCNN-Lite was 334 times smaller than its teacher, DiseasedCNN. The total size of the model, along with the shortened inference time, makes DiseasedCNN-Lite a more practical solution than DiseasedCNN, and opens the door to integration with agricultural robots and drones. Although a test accuracy of 87% is relatively low compared to other existing models (which routinely achieve accuracies above 90%), DiseasedCNN-Lite provides a strong foundation for future research into training smaller student models.
We believe that with slight changes to hyperparameters, and many more epochs of training, DiseasedCNN-Lite could increase its capabilities. We would have to be careful not to overfit to our dataset, and to prevent this our best option would likely be to increase our data augmentation. To best support an increase in DiseasedCNN-Lite’s abilities, we would continue to calibrate DiseasedCNN in order to optimize its accuracy. Although unlikely, it is possible that DiseasedCNN-Lite could achieve a higher test accuracy than its teacher, DiseasedCNN. However, a more accurate teacher model would only be more helpful during training.
Conclusions
DiseasedCNN-Lite achieved a respectable accuracy of 87%, while being an extremely lightweight model totaling only 390 kilobytes in size. Compared to the capabilities of its teacher, DiseasedCNN-Lite was capable of similar accuracies while shrinking the size of the model by a factor of 334. Overall, this model is still not ready to be deployed and tested with agricultural robots and drones, but it lays a solid foundation for future work.
One of the main limitations of DiseasedCNN-Lite is that it can only predict diseases given an image containing only one leaf. A potential solution to this problem is to use a vision transformer as a pretrained backbone, and teach DiseasedCNN-Lite to not only identify diseased leaves, but also locate leaves with bounding boxes if given an image with multiple leaves. We would also like to increase the test accuracy of DiseasedCNN-Lite, and perform tests on how well it generalizes to real-world data. The next step after those improvements is to deploy the model onto an agricultural robot or drone, and test its capabilities in the real world.
Citations
If you found our work helpful, consider citing us with the following BibTeX reference:
@article{2025DiseasedCNN-Lite,
title = {DiseasedCNN-Lite: Leveraging Transfer Learning and Knowledge Distallation for Leaf Disease Classification},
author = {Smith, David and Wu, Jess and Hasey, William},
year = {2025}
}
Contact
If you have any questions, feel free to contact any of David Smith, Jess Wu, or William Hasey.